Introduction
The eagle-i Research Resource Ontology (ERO) was developed in the context of the eagle-i project and funded through NIH/NCRR ARRA award #U24RR029825.
Goals
Our modeling approach had three main drivers. The first was to represent real data collected about resources. The second was to have the ontology control the user interface (UI) and the logic of the data collection tool and search application. The third was a commitment to build a set of ontologies that could be reusable and interoperable with other ontologies and existing efforts for representing biomedical entities.
This latter requirement translated into decisions to a) follow OBO Foundry principles and best practices for biomedical ontology development and b) engage in active discussions within the bio-ontology community in order to provide context for eagle-i interoperability and align with domain-wide standards for resource representation (http://bit.ly/rrcoord).
Content
We began our modeling effort by collecting a preliminary set of data with the goal of identifying key properties for each resource type collected by eagle-i. These include reagents, instruments, services, model and non-model organisms, protocols, biospecimens, human studies, and research opportunities.
We then asked the eagle-i team to identify a set of queries relevant to each of these resources. For example, “Which laboratories in the United States are equipped with high-resolution ultrasound machines for brachial artery reactivity testing (BART)?” or “Find in situ hybridization protocols for whole-mount preparations of Aplysia.”
Over 300 queries were generated and analyzed to first identify their relevance and semantic linkage, and then to specify the relations required to answer these queries. Using the preliminary data and the analysis of the queries, we defined a preliminary high-level data model. Next we identified a set of classes and properties that had to be created de novo, as well as those from existent biomedical ontologies that could be reused to implement this model. Currently, these include:
- Ontology for Biomedical Investigation (OBI)
- Basic Formal Ontology (BFO)
- RO (OBO Relation Ontology)
- Uber-Anatomy Ontology (Uberon)
- Ontology for Clinical Research (OCRe)
- Sequence Ontology (SO)
- Software Ontology (SWO)
- NCBI Taxonomy
- Disease Ontology (DO)
- Mammalian Phenotype Ontology (MP)
- PATO - Phenotypic Quality Ontology
- Gene Ontology (GO)
- Cell Line Ontology (CO)
- Information Artifact Ontology (IAO )
- VIVO Ontology
Implementation details
We used the Basic Formal Ontology (BFO) as upper ontology, the Information Artifact Ontology (IAO) for ontology metadata and the Relation Ontology (RO) for common relationships.
We applied the MIREOT principle in order to reference classes in existing ontologies.
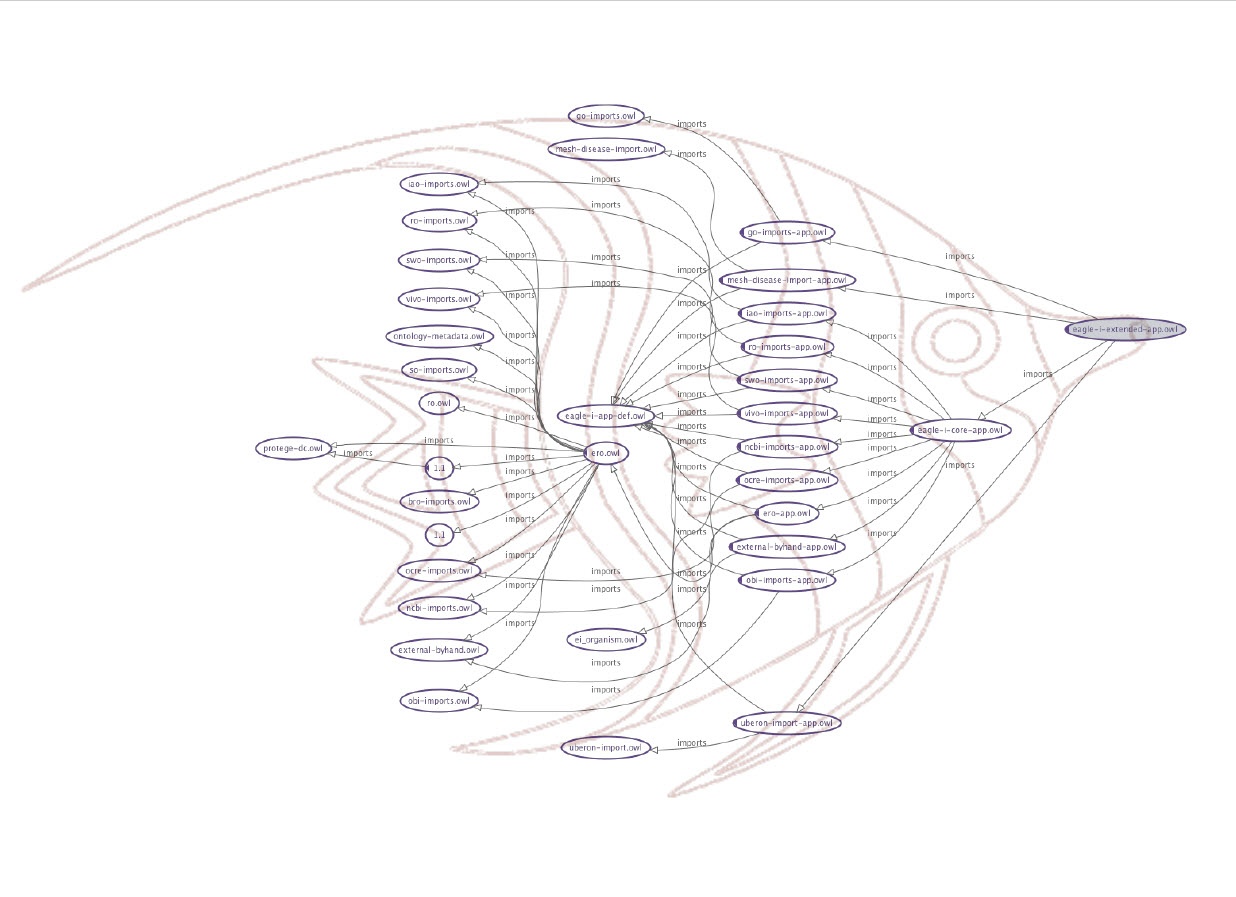
In order to be able to meet all the requirements we developed an import strategy to implement a layered approach that allows to decouple research resources representation from information used to drive application appearance and behavior. This import strategy is fondly referred to as the Messy FishTM (see Figure 1) or the ERO Matryoshka doll.
Figure 1
Lessons learned
In the process of developing ERO in compliance with eagle-i project requirements, we have learned important lessons related to best practices for biomedical ontology development and interoperability with other ontology-based resource systems and community ontologies :
1. Reuse of existing ontologies
Using upper level ontologies such BFO and trying to reuse existing ontologies following the OBO Foundry orthogonality principles has the advantage of guiding the development process and enabling the integration of other ontologies. However, when it comes to driving application UIs directly from an ontology, there is a need for implementation methods which exclude some upper level classes, specify or modify properties domain and range and deal with different modeling approaches (as not all the ontologies we have reused follow the same principles).
2. Use of layered import strategy
The strategy we used to separate the core and the application layer has shown its effectiveness as it has enabled parallel ontology development. The challenges it presents are related to keeping the annotations current and in preventing excessive proliferation of annotation properties as a quick way to simplify application development complexity. We have identified some common requirements in terms of annotations that are useful to drive a UI directly from an ontology and we will formalize them as design pattern.
3. Best practices and tools
For the development process, we have used Ontofox as a useful tool to MIREOT classes form other ontologies and generate our imports files. To drastically streamline this process, we developed scripts to automate the syncing of import files. To have the whole MIREOT roundtrip process integrated into an ontology editor such Protege would be ideal (i.e. selection of terms from an external ontology, updates to imported terms in the proper import files and checks of the ontology's consistency). We also identified the need to provide some community views or 'slims' with different content granularity and axiomatic complexity. Although at the present time there are some useful tools to facilitate this process (Ontodog) to have these facilities integrated into an editing tool would be ideal.
Review process
Before the official release we have asked Jie Zheng and Melanie Courtot to review the ontology. The reviewers' comments and the actions taken are available in this Google doc.
For more info about our development strategies and lessons learned check our publications.