The toolkit described herein is currently not user-friendly (though it works well – we use it routinely to bulk-upload data for the Consortium members). If you encounter issues, please do not hesitate to contact us.
SWIFT (Semantic Web Ingest from Tables) is a toolkit that allows experienced users to bulk-upload data into an eagle-i repository, via ETL (Extract, Transform and Load). The figure below is a high level depiction of the ETL process. Currently the toolkit supports only Excel spreadsheets as input files.
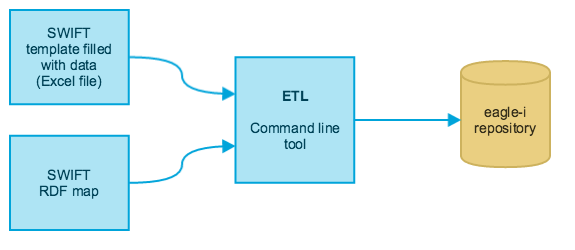
This guide provides an overview of tasks pertaining to ETL and the usage of the SWIFT toolkit. The ETL workflow requires a person with domain knowledge and understanding of the eagle-i resource ontology to prepare the input files for optimal upload, and a person with basic knowledge of Unix to run the commands and troubleshoot potential errors. A detailed description is provided in: Data preparation and ETL Workflow
The SWIFT toolkit is comprised of:
- an ETL Input Generator - command line program that generates spreadsheet templates and mapping files for the various resource types of the eagle-i resource ontology (e.g. a template/map for antibodies, for instruments, etc.)
- an ETLer - command line program that executes a bulk upload
- a deETLer - command line program that deletes a previous ETL upload
- Bulk workflow - command line program that executes workflow transitions on groups of resources, e.g. Publish, Return to curation, Withdraw
Prerequistes
The SWIFT Toolkit requires:
- A Unix-like environment including a terminal for executing commands
- MacOS and Linux users don't need to install anything extra. For MacOS, use the Terminal app under Applications/Utilities
- cygwin is recommended for Windows users
- A Java 1.7 runtime environment
- execute the command {{ java -- version}} to find out what version you have.
- If necessary, you may download the JRE directly from Oracle and follow the installation instructions.
Download
The SWIFT toolkit is packaged as a zip file, and can be downloaded from our software repository.
Download the SWIFT toolkit distribution that matches the version of your eagle-i repository, named eagle-i-datatools-swift-[version]-dist.zip, Unzip it into a dedicated directory, and navigate to it. For example
mkdir ~/eagle-i unzip -d ~/eagle-i eagle-i-datatools-swift-2.0MS3.01-dist.zip cd ~/eagle-i/swift-2.0MS3.01
Available commands
Input generation
To generate etl templates and maps, navigate to the dedicated directory (above) and run the script:
./generate-inputs.sh -t typeURI
*You may obtain the type URI from the eagle-i ontology browser . Use the left bar to find the most specific type you need, select it and grab its URI, e.g. http://purl.obolibrary.org/obo/ERO_0000229 for Monoclonal Antibodies.
Innocuous warnings are produced when generating the templates; these may safely be ignored. If you encounter errors or issues, please do not hesitate to contact us.
This script will create/use two directories with obvious meanings: ./maps and ./templates.
ETL
The ETLer expects data to be entered into one of the generated templates, and a few conventions to be respected (see Data preparation and ETL Workflow) . A data curator usually makes sure that the template is correctly filled. In particular, the location of the resources to be ETLd (e.g. Lab or Core facility name) must be provided in every row of data.
- Place your input files (i.e. the completed templates) in a directory of your choice, e.g.
dataDirectory. All files contained in this directory will be processed by the ETLer. To run an ETL, execute the following command. Note that all resources will be uploaded in the requested workflow state - we recommend to choose CURATION, verify the resources were ETLd correctly, and then publish using the bulk workflow command (see below). If you've already ran a test ETL in a staging environment, choose PUBLISH directly.
./ETLer.sh -d dataDirectory [-p DRAFT|CURATION|PUBLISH] -c username:password -r repositoryURL
If you are practicing the ETL process, you may wish to upload your data to the common eagle-i training node. In this case, if your directory is named dataDirectory, the script would be executed as follows (default workflow state is DRAFT):
./ETLer.sh -d dataDirectory -c L4:Level4 -r https://training.eagle-i.net
Note that the data that is uploaded to the training node CAN be viewed and modified by others even in a draft state (even if you subsequently lock the records). Note also that the information in the training node is not persistent as the node is refreshed periodically.
- A detailed report of the ETL results is generated in the
./logsdirectory; please inspect it to verify that all rows were correctly uploaded. The RDF version of generated resources is also logged in this directory. - To further verify the data upload, log on to the SWEET application and select the lab to which the ETLd resources belong.
De-ETL
Resources that are uploaded to an eagle-i repository via ETL are tagged with the name of the file from which they were extracted. It is therefore relatively simple to de-ETL an entire file. To do so, execute the following command:
./deETLer -f filename -c username:password -r repositoryURL
Bulk Workflow
Execute the following command to perform workflow actions (e.g. send to curation, publish, unpublish) on all resources ETLd from a particular file (i.e. resources that are tagged with the filename in the eagle-i repository):
./bulk-workflow -f filename -p DRAFT|CURATION|PUBLISH -c username:password -r repositoryURL
Note the following limitations of bulk workflow:
- All the resources tagged with the filename must be in the same state
- You must choose a final state that is reachable from the resources' current state
- if the resources are in draft, choose CURATION
- if the resources are in curation, choose PUBLISH or DRAFT
- if the resources are published, choose CURATION
- if you want to publish resources that are currently in draft, you'll need to run the bulk workflow command twice